Antimicrobial resistance surveillance
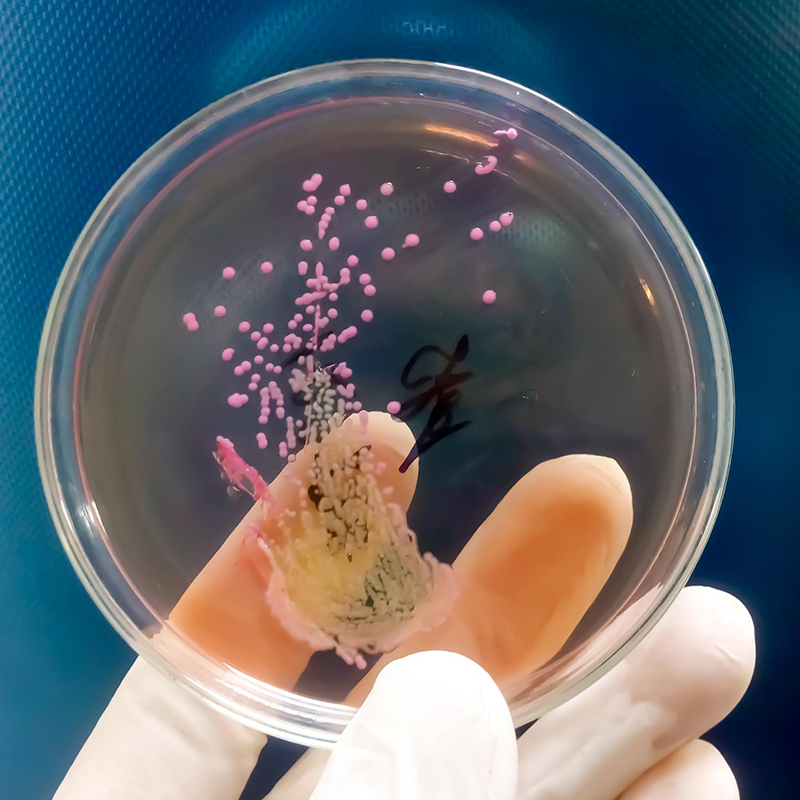
What is antimicrobial resistance (AMR) detection?
Antimicrobial resistance (AMR) detection is the process of identifying microorganisms (such as bacteria, viruses, fungi, or parasites) that are resistant to antimicrobial drugs commonly used to treat infections.1,2 A goal of AMR detection is to provide information about the resistance of pathogens to different antimicrobial drugs and to guide decisions to minimize the spread of resistant strains.3
AMR is primarily based on the ability of microorganisms to adapt through genomic changes. These genomic changes can be detected using the power of next-generation sequencing (NGS), which offers unprecedented high-throughput capabilities to identify low-frequency variants and genomic arrangements.
How is antimicrobial resistance detection used?
When coupled with the power of NGS, AMR can be efficiently used for:
Early detection
Stay ahead of emerging resistance markers through resistome monitoring. See how scientists are using early detection to minimize spread of AMR pathogens.
Detecting a resistance gene in Staphylococcus sciuri
This paper reports the discovery of a resistance gene found in a novel mosaic plasmid, highlighting the need to implement genome-wide studies to reveal the identification, characterization, and evolution of AMR bacteria.
Urban monitoring of AMR
Scientists show how wastewater analysis can be used along with traditional surveillance efforts to help identify current and emerging AMR threats.
Outbreak responses
Tracking transmission routes and monitoring variants of concern are vital areas of focus during an outbreak response. Read how scientists are dealing with outbreaks with NGS-based detection methods.
Using genomics for global AMR tracking
This publication provides an overview of how AMR tracking can benefit from NGS-based solutions and applications such as WGS to enhance real-time data during outbreaks.
Detecting horizontal gene transfer in multidrug-resistant bacteria
In this article, scientists screened bacterial genomes from healthcare-associated infections from a single hospital and showed that transfer of genetic elements, including those that provided resistance to several drugs, occurred in this setting.
Clinical research
Insights into disease mechanisms not only enable the development of diagnostics and therapeutics but also allow strategic responses that could prevent further AMR resistance. See the papers below to learn more about how advancements in AMR detection using NGS can be used to gain an edge in controlling pathogen resistance.
Acquiring AMR in travel
Learn how investigators combine NGS, functional metagenomics, and statistical modeling to study the abundance, diversity, function, context, and acquisition of AMR genes.
Sequencing-based methods to study AMR
This informative review covers antimicrobial resistance identification and characterization methods, including recent deep-learning methods. In addition, the authors discuss sequencing-based resistance discovery along with tools and databases used in antimicrobial resistance studies.
Detecting AMR from WGS data
Learn more about how researchers designed and validated their bioinformatic tool to detect genomic determinants of AMR in bacteria from WGS data.
NGS-based methods to detect AMR
NGS is a revolutionary tool offering breakthrough detection capabilities in the fight against AMR. Illumina offers innovative solutions from sample to data to keep pace with the rapid ability of microbes to evolve against antimicrobial compounds. Explore our expanding NGS-based solutions to make your next advancement in AMR detection.
Whole-genome sequencing (microbial)
With whole-genome sequencing (WGS), you can obtain comprehensive genomic insights into microbial origins, identification, phenotype, evolutionary behavior, and more that are critical for AMR-related studies. Take advantage of high-resolution variant detection without the need for specific probes. Powered by NGS, microbial WGS can be performed with simplified workflows to sequence many samples through the power of multiplexing.
Shotgun metagenomics
Targeting genes within complex samples to study emerging diseases or activity related to AMR is possible with shotgun metagenomics. This method allows you to collectively sample all genes from all organisms, including those that are difficult to culture without the need for target enrichment. As an NGS-based application, shotgun metagenomics enables high sequence coverage to detect low-abundance, antimicrobial-resistant microbes while being able to analyze thousands of samples in parallel.
Hybrid capture
When only certain genes or specific genomic regions need to be sequenced, targeted enrichment methods allow you to efficiently focus efforts and resources. Targeted enrichment allows you to detect AMR sensitively and accurately, for example, by identifying novel variants with high mutation resolution.
Interested in receiving the latest updates about microbiology research?
Featured Products
Respiratory Pathogen ID/AMR Enrichment Panel
This NGS target enrichment workflow identifies a broad range of respiratory pathogens and antimicrobial resistance alleles, and offers simplified data analysis powered by IDbyDNA.
Order Now
AmpliSeq for Illumina Antimicrobial Resistance Panel
Target 478 AMR genes to evaluate antibiotic treatment efficacy for 28 antibiotic classes.

Illumina DNA Prep
The Illumina DNA Prep is a fast, user-friendly solution for a wide range of applications such as microbial species. This kit is also designed to allow DNA extraction from diverse sample types with the flexibility to sequence large, complex genomes as well as amplicons or microbial species.

Urinary Pathogen ID/AMR Enrichment Kit
The Urinary Pathogen ID/AMR Panel (UPIP) is designed to identify uropathogens responsible for urinary tract infections (UTIs) and co-infections, detect AMR, and perform strain typing of critical pathogens to study evolution and transmission.
Related resources

Public health surveillance
Learn how public health surveillance can be a vital tool for identifying infectious disease pathogens, variants, AMR and more.
Healthcare-associated infection surveillance
Get an overview of healthcare-associated infection surveillance and related solutions from Illumina.

Wastewater surveillance
Explore how wastewater surveillance can identify pathogens, including strains related to AMR, and variants in a community.

Microbial whole-genome sequencing
See Illumina solutions for microbial whole-genome sequencing to analyze hundreds of organisms with the power of multiplexing. Confidently detect low-frequency variants and genome rearrangements.

Microbiology sequencing platform comparison tool
Illumina offers a wide rage of platforms to enable your next discovery in AMR. Use this platform to compare NGS systems to find the one that is right for your lab or application.
Transforming sequencing data into meaningful results
NGS produces massive amounts of data that requires analysis using bioinformatics. Explore analysis solutions that decipher sequencing data into AMR-related insights.
References
- About Antibiotic Resistance. cdc.gov/drugresistance/about. Accessed September 20, 2023.
- Antimicrobial resistance. who.int/news-room/fact-sheets/detail/antimicrobial-resistance. Accessed September 20, 2023.
- Antimicrobial Resistance Information from FDA. fda.gov/emergency-preparedness-and-response/mcm-issues/antimicrobial-resistance-information-fda. Accessed September 20, 2023.