Microbial Whole-Genome Sequencing
Introduction to Microbial Whole-Genome Sequencing
Microbial whole-genome sequencing is an important tool for mapping genomes of novel organisms, finishing genomes of known organisms, or comparing genomes across multiple samples. Sequencing entire bacterial, viral, and other microbial genomes is important for generating accurate reference genomes, for microbial identification, and for other comparative genomic studies.
Unlike capillary sequencing or PCR-based approaches, next-generation sequencing (NGS) allows microbiology researchers to sequence hundreds of organisms with the power of multiplexing. Unlike traditional methods, NGS-based microbial genome sequencing doesn’t rely on labor-intensive cloning steps, saving time and simplifying the workflow. NGS can identify low-frequency variants and genome rearrangements that may be missed or are too expensive to identify using other methods.

Methods Guide for Microbial Whole-Genome Sequencing
Sequence hundreds of organisms simultaneously, generate accurate reference genomes, and perform comparative genomic studies.
Download GuideDe Novo Microbial Genome Sequencing
De novo whole-genome sequencing involves assembling a genome without the use of a genomic reference and is often used to sequence novel microbial genomes. Illumina sequencers provide unparalleled raw read accuracy, read length and read depth for high-quality draft and complete microbial genome assemblies.
Microbial Whole-Genome Resequencing
Microbial whole-genome resequencing involves sequencing the entire genome of a bacteria, virus, or other microbe, and comparing the sequence to that of a known reference. Generating rapid and accurate microbial genome sequence information is critical for detecting low frequency mutations, finding key deletions and insertions, and discovering other genetic changes among microbial strains.
Featured Microbial Whole-Genome Sequencing Workflow
This next-generation sequencing workflow is designed for beginners and describes the recommended methods and products for each step of this application.
View Workflow
Featured Products
Featured Microbial Research Articles
Illumina Sequencers Help Characterize and Control Coronavirus
See how Illumina NGS was used to sequence the novel coronavirus genome, in combination with other technologies, as published in the New England Journal of Medicine.
Read Article
The Fight Against Ebola
In 2014, the world's eyes were pointed toward West Africa, where an Ebola outbreak was claiming the lives of thousands of people. Long before that, the virus was quietly circulating.
Read Article
The Power and Portability of the iSeq
Local scientists used the iSeq 100 System to analyze transmission patterns and trace the origin of the Ebola outbreak in the Democratic Republic of Congo.
Read ArticleMicrobiology Methods Guide
All the information you need, from library preparation to final data analysis. Select the best tools for a broad range of microbiology applications for your laboratory.
Access Guide
Suggested Workflow for Microbial Whole-Genome Sequencing
There are multiple ways to perform these experiments, but these are some suggested products for each step of the workflow.
Click on the below to view products for each workflow step.
A fast, integrated workflow for a wide range of applications, from human whole-genome sequencing to amplicons, plasmids, and microbial species.
Nextera XT Library Prep KitPrepare sequencing-ready libraries for small genomes (bacteria, archaea, viruses), amplicons, and plasmids in less than 90 minutes.
Simple, all-inclusive library preparation with shortened gel-free workflows, the ability to sequence challenging regions, and the power to identify variants across the genome.
Affordable, fast, and accessible sequencing power for targeted or small genome sequencing in any lab.
MiniSeq SystemSupports a broad range of targeted DNA and RNA applications.
MiSeq SystemSeveral kit configurations, including a 500 cycle kit for efficient mapping of small genomes. Micro and nano formats available for cost-efficient sequencing of 1-4 samples.
Groundbreaking benchtop sequencers allow you to explore new discoveries across a variety of current and emerging applications.
NovaSeq 6000 SystemScalable throughput and flexibility for virtually any genome, sequencing method, and scale of project.
BaseSpace Apps for de novo assemblies or mapping of contigs and scaffolds
Velvet de novo AssemblyDe novo assembly of bacteria using the Velvet assembler with a focus on Nextera Mate Pair data.
String Graph AssemblerPerforms a contig assembly, builds scaffolds, removes mate pair adapter sequences, and calculates assembly quality metrics.
SPAdes Genome AssemblerOnline bioinformatics tool for assembling microbial genome sequences.
Rapidly annotates genes and identifies coding sequences in prokaryotic genomes, from de novo assembly sequences.
RescafImproves scaffolding in bacterial de novo assemblies by removing assembly errors and closing gaps in the consensus sequence.
TELLSeqTell-Read and Tell-Link App links reads and creates assemblies for de novo WGS.
Frequently Purchased Together
Interested in receiving newsletters, case studies, and information on microbial genomics? Enter your email address.
Additional Resources
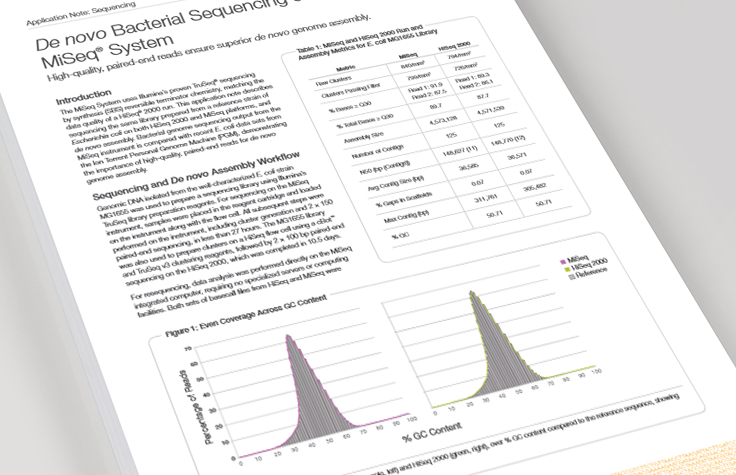
De Novo Bacterial Sequencing Overview
Learn how high-quality paired-end reads ensure superior bacterial genome assembly.

Microbiology: Spotlight on Food Pathogen Testing
Overview of Illumina next-generation sequencing platforms and relevant applications and use cases for food pathogen testing.
Push-Button De Novo Microbial Genome Sequence Assembly
Explore options for performing de novo assembly of microbial and other genomes.

Assembly of the Saccharomyces cerevisiae Genome
This application note describes a complete workflow for sequencing small genomes with MiSeq.

NGS for Microbial Genomes and Infection Control
Global collaboration of researchers is using the MiSeq System to understand the evolution and spread of drug-resistant bacteria.

TELL-Seq: Bringing Speed and Accuracy to Long-Range Sequencing
TELL-Seq technology is a simple library prep solution which enables Illumina NGS systems to generate highly accurate and economical long-range sequencing information.
References
- Köser CU, Bryant JM, Becq J, Török ME, Ellington MJ, et al. (2013) Whole-genome sequencing for rapid susceptibility testing of M. tuberculosis. N Engl J Med 369(3):290–2.
- Harrison EM, Paterson GK, Holden MT, Larsen J, Stegger M, et al. (2013) Whole-genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol Med 5(4):509–15.
- Kato-Maeda M, Ho C, Passarelli B, Banaei N, Grinsdale et al. (2013) Use of whole genome sequencing to determine the microevolution of Mycobacterium tuberculosis during an outbreak. PLoS One 8(3):e58235.