Targeted Resequencing
Introduction to Targeted Resequencing
With targeted resequencing, a subset of genes or regions of the genome are isolated and sequenced. Targeted approaches using next-generation sequencing (NGS) allow researchers to focus time, expenses, and data analysis on specific areas of interest. Such targeted analysis can include the exome (the protein-coding portion of the genome), specific genes of interest (custom content), targets within genes, or mitochondrial DNA.
Benefits of Targeted Resequencing
Targeted resequencing is simpler and more accessible than ever before. Find out how to access the focused power of targeted NGS.
Affordable Sequencing With Focused Content
- Focuses on regions of interest, generating a smaller, more manageable data set
- Reduces sequencing costs and data analysis burdens
- Reduces turnaround time compared to broader approaches
- Enables deep sequencing at high coverage levels for rare variant identification
Compared to broader approaches, such as whole-genome sequencing, targeted sequencing is a more cost-effective method for investigating areas of interest. Targeted resequencing enables researchers to focus on regions that are most likely to be involved in the phenotype under study, conserving resources and generating a smaller, more manageable data set. Targeted approaches can also deliver much higher coverage levels, allowing identification of variants that are rare and more expensive with whole-genome or Sanger sequencing.
Using qPCR or Sanger sequencing today?
Find out how Illumina targeted sequencing can help you discover more.

How can I use NGS to analyze specific genes or DNA regions?
Explore common targeted NGS methods, from exome to amplicon sequencing and more.
How can I apply targeted NGS?
Discover, validate, or screen genetic variants with targeted sequencing for various research areas, from cancer to microbiology and more.
Explore uses of targeted NGSFeatured Targeted Sequencing Workflow: Custom Amplicon Sequencing
This key method uses ultra-deep sequencing of PCR amplicons to target specific genomic regions.
Content Selection
DesignStudio Assay Designer
Online tool makes it easy to optimize for target region coverage.
AmpliSeq for Illumina Custom DNA Panel
Create custom targeted sequencing panels optimized for content of interest.
Library Prep & Accessories
AmpliSeq for Illumina Library PLUS, Indexes & Accessories
Rapidly prepare amplicon libraries for Illumina sequencers.
Sequencing
iSeq 100 System
Affordable, fast, and accessible sequencing power for targeted or small genome sequencing in any lab.
MiniSeq System
Supports a broad range of targeted DNA and RNA applications.
Data Analysis
Local Run Manager (DNA Amplicon Analysis Module)
An on-premises software solution for creating sequencing runs, monitoring run status, and analyzing data.
BaseSpace Sequence Hub (DNA Amplicon App)
The Illumina genomics cloud computing environment for NGS data analysis and management.
Identify and Track Infectious Disease Threats
NGS supports effective genomic surveillance strategies to reduce infectious disease transmission. Targeted sequencing can be used to monitor zoonotic or respiratory pathogens.
Learn More About Genomic Surveillance
Interested in receiving newsletters, case studies, and information on genomic analysis techniques? Enter your email address.
Additional Resources

Advancing Clinical Cancer Research
Targeted NGS enables cancer researchers to cost-effectively assess multiple genes in a single test.
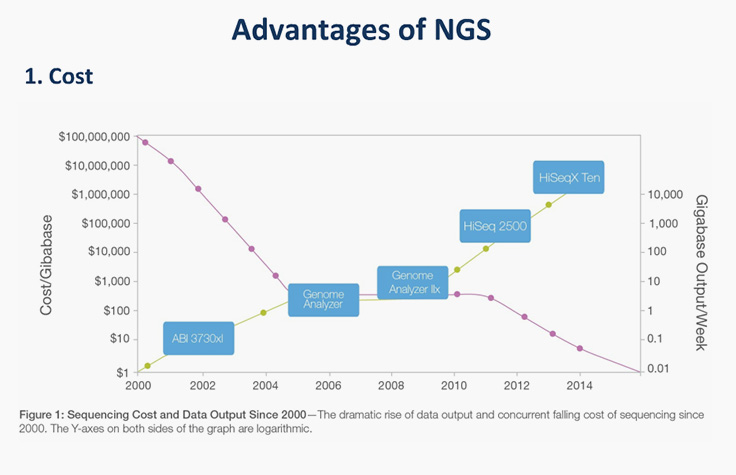
Basics of NGS for FFPE Tumor Specimens
Learn how to start using targeted NGS in your clinical research lab.
Targeted Sequencing for HIV Studies
Researchers used targeted resequencing to detect minor variants associated with human immunodeficiency virus (HIV) tropism.

Transitioning to AmpliSeq for Illumina
Our detailed guide is designed to help you transfer projects from TruSeq Custom Amplicon to AmpliSeq for Illumina panels.

AmpliSeq for Illumina on the iSeq 100 System
Find step-by-step guidance designed to help you transition your workflows to AmpliSeq for Illumina panels on the iSeq 100 System.